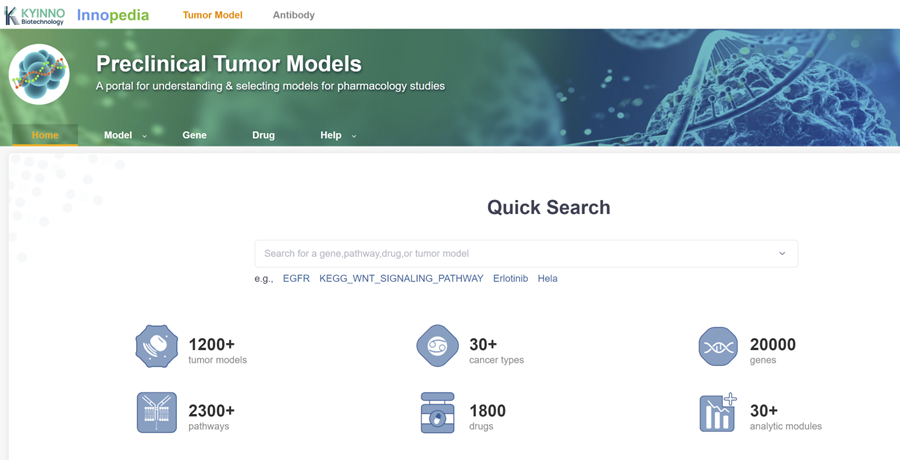
Innopedia Database
Innopedia - Embark on a Fresh Journey into Cell Line Exploration!
Kyinno’s cutting-edge Innopedia database is now officially launched!
Register and use it for free: https://innopedia.kyinno.com
Innopedia is a robust database designed to provide researchers with enhanced research tools and data support. The database offers browsing, searching, and analytical capabilities for tumor cell lines, engineered cell lines, and standard antibodies. It encompasses over 600 commonly used tumor cell lines, 700 engineered cell lines with essential genomic and pharmacological data, cell line efficacy data for nearly 2000 common anti-tumor drugs, and comprehensive details for over 100 standard antibodies. Researchers can conveniently and precisely choose the most suitable cell lines for pharmacological and efficacy experiments using this database.
Innopedia boasts over 30 analysis modules that empower researchers to effectively analyze and comprehend data. For instance, researchers can utilize the “Gene Expression” module to examine gene expression profiles across different cell lines, facilitating the selection of cell lines suited to their research. The “Pharmacological Experiment” module allows the evaluation of the sensitivity of various cell lines to specific drugs, aiding in the selection of appropriate cell lines for pharmacological experiments. The “Gene Variation” module assists in identifying tumor-related gene variations within different cell lines.
Furthermore, Innopedia provides practical functionalities such as “Cell Line Comparison” to contrast distinctions between various cell lines and “Drug Screening” to filter drugs relevant to specific diseases.
In summary, Innopedia is a potent tool that helps researchers gain a better understanding of data in the fields of tumor and drug research, ultimately advancing scientific investigations. We believe that the release of this database will have a positive impact on the realms of tumor and drug research.
Here are some examples illustrating the database's functions:
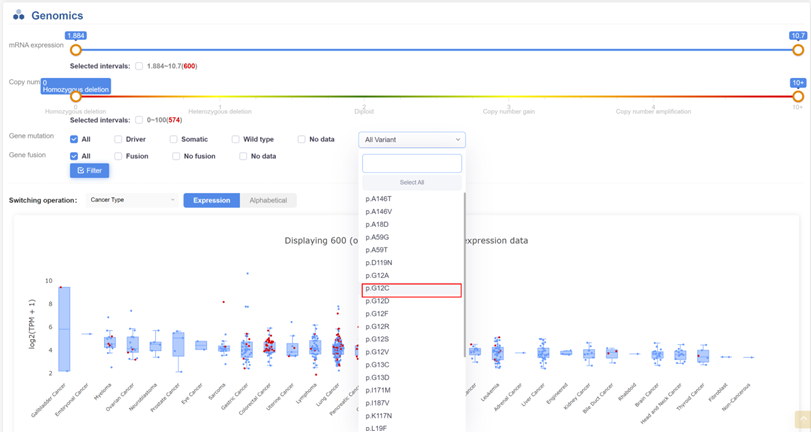
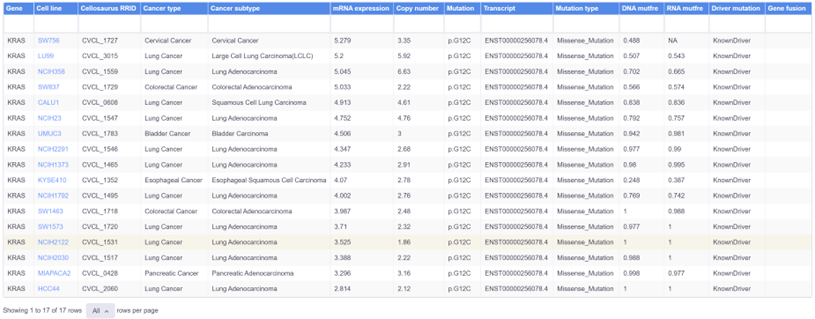
1. Retrieving Cell Lines with KRAS-G12C Mutation:
Navigate to the KRAS gene page.
Select the p.G12C mutation from the available options.
2. Retrieving Cell Lines with High or Low EGFR Expression:
Go to the EGFR gene page.
Choose EGFR expression values >7 or <0.1.
This will yield 22 cell lines with high expression and 37 cell lines with low expression. Among them, 8 cell lines (highlighted in red) carry EGFR mutations.

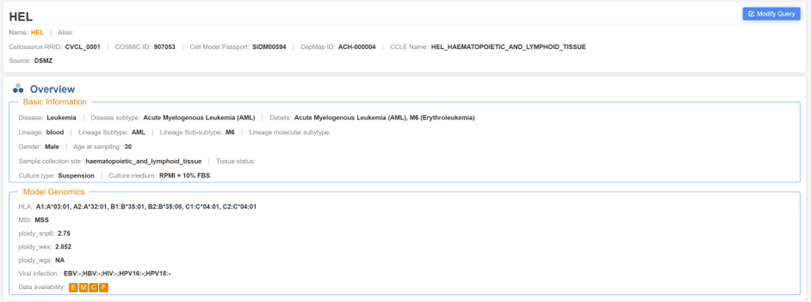
3. Retrieving HEL cell line information

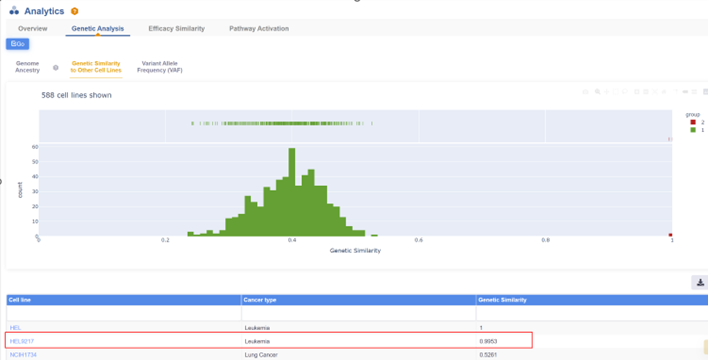
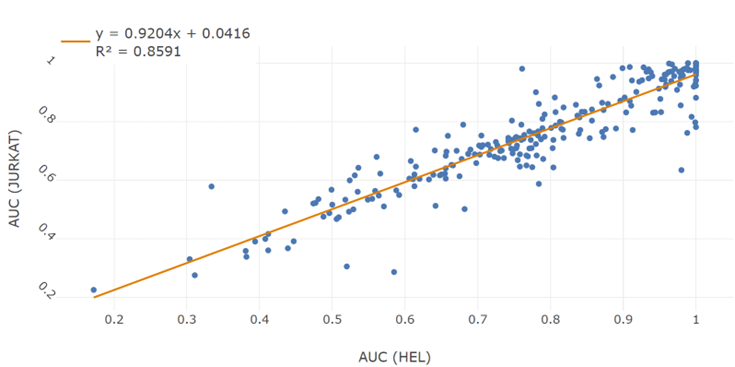
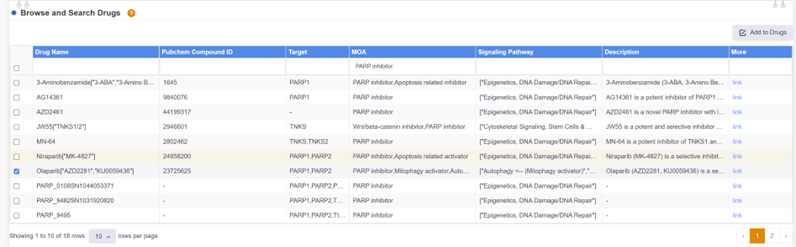
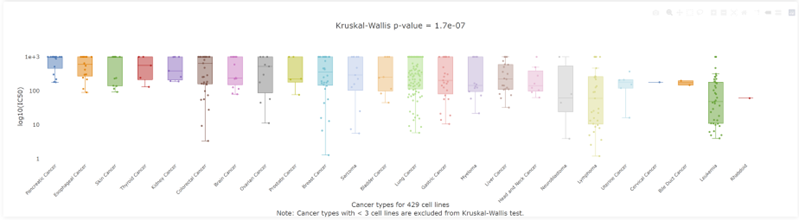
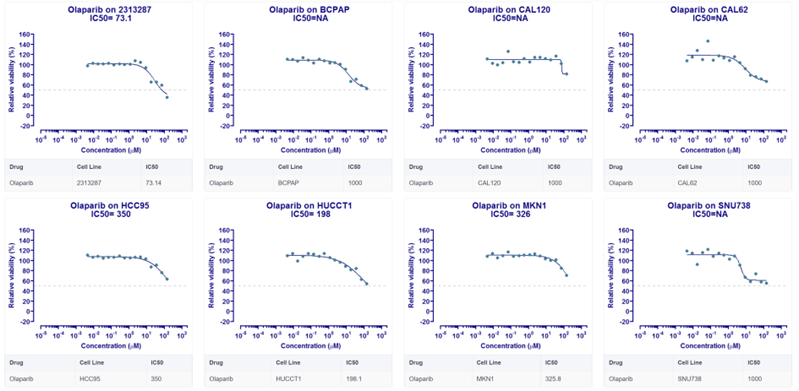
4. Retrieving the efficacy trial data of PARP inhibitor Olaparib
5. Retrieving engineered cell lines carrying EML4-ALK gene fusion

6. Retrieving standard antibody information

